from IPython.display import Image
from IPython.core.display import HTML
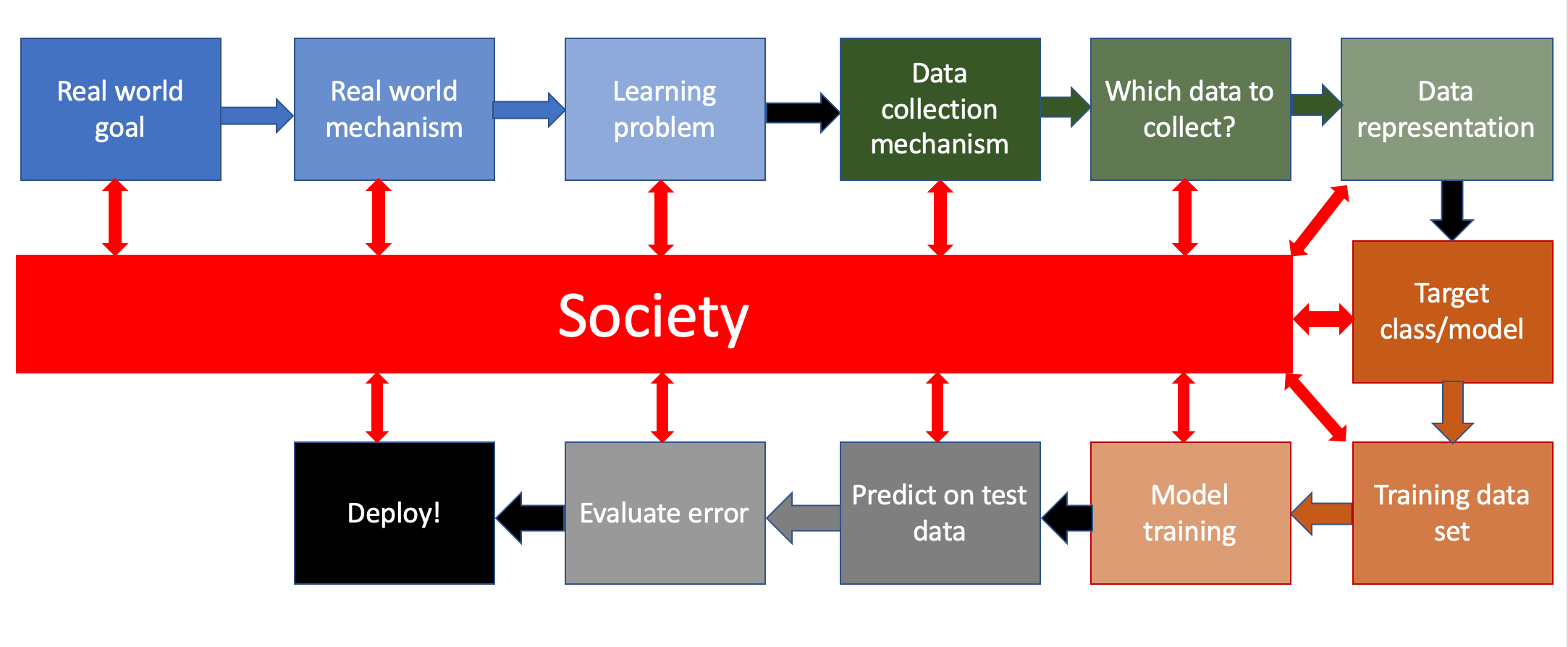
Image(url= "http://www-student.cse.buffalo.edu/~atri/algo-and-society/support/notes/pipeline/images/ML-pipeline-duame-soc.png",
width=800)
import numpy as np
import pandas as pd
import plotly.express as px
We're going to take a look at data on COVID coverage by local news agencies in the US.
There are a number of features in this dataset, there's a more detailed description of most of them here. I'll describe what we're using here, though, obviusly
Note: I am, again, using data that I have used for research in the past. I think it is useful, because I am able to provide more thoughtful responses to questions and comments about the data and methods. It is not because I think my research is especially great, or that you should read it.
covid_df = pd.read_csv("covid_local_news.csv")
from IPython.display import Image
from IPython.core.display import HTML
Image(url= "http://www-student.cse.buffalo.edu/~atri/algo-and-society/support/notes/pipeline/images/ML-pipeline-duame-soc.png",
width=800)

Image(url="https://pbs.twimg.com/media/FKtEyl5XEAABdEa?format=jpg&name=4096x4096", width=800)

Find local news deserts. In other words, find places where people aren('t) likely to be getting adequate news about COVID
Identify places where the number of articles that cover COVID is low. We can then try to forecast coverage in locations where we don't have data.
from IPython.display import Image
from IPython.core.display import HTML
Image(url= "./coverage.png", width=600, height=600)

px.histogram(covid_df, x="n_covid_full_filter")
px.histogram(covid_df, x="perc_full")
covid_df['log_odds_of_covid_article'] = np.log((covid_df.n_covid_full_filter+1)/(covid_df.n_total_articles-covid_df.n_covid_full_filter+1))
px.histogram(covid_df, x="log_odds_of_covid_article")
Predict the percentage of weekly coverage devoted to COVID for local news outlets across the country that make data available
pd.set_option('display.max_columns', None)
covid_df.head()
| fips_full | fips | state | week_num | fips_covid | sourcedomain_id | lon | lat | title | rank | n_total_articles | n_covid_limited_filter | n_covid_full_filter | fips_cum_cases | fips_cum_deaths | fips_lag1_cum_cases | fips_lag2_cum_cases | fips_n_cases | fips_lag1_n_cases | fips_lag1_cum_deaths | fips_lag2_cum_deaths | fips_n_deaths | fips_lag1_n_deaths | state_cum_cases | state_cum_deaths | state_lag1_cum_cases | state_lag2_cum_cases | state_n_cases | state_lag1_n_cases | state_lag1_cum_deaths | state_lag2_cum_deaths | state_n_deaths | state_lag1_n_deaths | country | country_cum_cases | country_cum_deaths | country_lag1_cum_cases | country_lag2_cum_cases | country_n_cases | country_lag1_n_cases | country_lag1_cum_deaths | country_lag2_cum_deaths | country_n_deaths | country_lag1_n_deaths | date | county | trump16 | clinton16 | total_population | white_pct | black_pct | hispanic_pct | age65andolder_pct | lesshs_pct | rural_pct | ruralurban_cc | state_popn | POPESTIMATE2019 | country_population | lo_trump_vote_16 | fips_n_cases_per1k | fips_n_cases_per1k_log | fips_n_cases_lo | fips_lag1_n_cases_per1k | fips_lag1_n_cases_per1k_log | fips_lag1_n_cases_lo | fips_n_deaths_per1k | fips_n_deaths_per1k_log | fips_n_deaths_lo | fips_lag1_n_deaths_per1k | fips_lag1_n_deaths_per1k_log | fips_lag1_n_deaths_lo | state_n_cases_per1k | state_n_cases_per1k_log | state_n_cases_lo | state_lag1_n_cases_per1k | state_lag1_n_cases_per1k_log | state_lag1_n_cases_lo | state_n_deaths_per1k | state_n_deaths_per1k_log | state_n_deaths_lo | state_lag1_n_deaths_per1k | state_lag1_n_deaths_per1k_log | state_lag1_n_deaths_lo | country_n_cases_per1k | country_n_cases_per1k_log | country_n_cases_lo | country_lag1_n_cases_per1k | country_lag1_n_cases_per1k_log | country_lag1_n_cases_lo | country_n_deaths_per1k | country_n_deaths_per1k_log | country_n_deaths_lo | country_lag1_n_deaths_per1k | country_lag1_n_deaths_per1k_log | country_lag1_n_deaths_lo | perc_full | perc_lim | predrt_0 | predrt_3 | predrt_12 | popuni | log_pop | log_rank | fips5 | Biden | Trump | lo_trump_vote | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1013 | 1013 | Alabama | 14 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 4 | 2 | 4 | 8 | 0 | 2 | 1 | 6 | 1 | 0 | 0 | 0 | 0 | 3955 | 115 | 2200 | 1008 | 1755 | 1192 | 64 | 14 | 51 | 50 | USA | 604363 | 29145 | 393848 | 182876 | 210515 | 210972 | 14501 | 3974 | 14644 | 10527 | 2020-04-08 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 30.851501 | 3.461085 | -7.929641 | 5.141917 | 1.815137 | -9.182404 | 0.000000 | 0.000000 | -9.875551 | 0.000000 | 0.000000 | -9.875551 | 17.896531 | 2.938978 | -8.627749 | 12.155364 | 2.576830 | -9.014316 | 0.520070 | 0.418756 | -12.147299 | 0.509873 | 0.412025 | -12.166717 | 32.067284 | 3.498544 | -8.045084 | 32.136898 | 3.500647 | -8.042916 | 2.230688 | 1.172695 | -10.710547 | 1.603555 | 0.956878 | -11.040608 | 1.000000 | 1.000000 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 |
| 1 | 1013 | 1013 | Alabama | 15 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 11 | 1 | 7 | 16 | 0 | 8 | 2 | 8 | 6 | 0 | 0 | 0 | 0 | 5350 | 195 | 3955 | 2200 | 1395 | 1755 | 115 | 64 | 80 | 51 | USA | 804845 | 44373 | 604363 | 393848 | 200482 | 210515 | 29145 | 14501 | 15228 | 14644 | 2020-04-15 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 41.135335 | 3.740887 | -7.678326 | 30.851501 | 3.461085 | -7.929641 | 0.000000 | 0.000000 | -9.875551 | 0.000000 | 0.000000 | -9.875551 | 14.225447 | 2.722968 | -8.857177 | 17.896531 | 2.938978 | -8.627749 | 0.815796 | 0.596524 | -11.704094 | 0.520070 | 0.418756 | -12.147299 | 30.538979 | 3.451224 | -8.093917 | 32.067284 | 3.498544 | -8.045084 | 2.319648 | 1.199859 | -10.671445 | 2.230688 | 1.172695 | -10.710547 | 0.636364 | 0.636364 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 |
| 2 | 1013 | 1013 | Alabama | 16 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 1 | 0 | 1 | 45 | 1 | 16 | 8 | 29 | 8 | 0 | 0 | 1 | 0 | 6752 | 245 | 5350 | 3955 | 1402 | 1395 | 195 | 115 | 50 | 80 | USA | 1012453 | 58064 | 804845 | 604363 | 207608 | 200482 | 44373 | 29145 | 13691 | 15228 | 2020-04-22 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 149.115590 | 5.011406 | -6.474354 | 41.135335 | 3.740887 | -7.678326 | 5.141917 | 1.815137 | -9.182404 | 0.000000 | 0.000000 | -9.875551 | 14.296830 | 2.727646 | -8.852175 | 14.225447 | 2.722968 | -8.857177 | 0.509873 | 0.412025 | -12.166717 | 0.815796 | 0.596524 | -11.704094 | 31.624467 | 3.485063 | -8.058990 | 30.538979 | 3.451224 | -8.093917 | 2.085520 | 1.126720 | -10.777834 | 2.319648 | 1.199859 | -10.671445 | 1.000000 | 1.000000 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 |
| 3 | 1013 | 1013 | Alabama | 17 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 12 | 3 | 7 | 120 | 2 | 45 | 16 | 75 | 29 | 1 | 0 | 1 | 1 | 8438 | 317 | 6752 | 5350 | 1686 | 1402 | 245 | 195 | 72 | 50 | USA | 1203928 | 70834 | 1012453 | 804845 | 191475 | 207608 | 58064 | 44373 | 12770 | 13691 | 2020-04-29 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 385.643768 | 5.957504 | -5.544818 | 149.115590 | 5.011406 | -6.474354 | 5.141917 | 1.815137 | -9.182404 | 5.141917 | 1.815137 | -9.182404 | 17.192906 | 2.901032 | -8.667836 | 14.296830 | 2.727646 | -8.852175 | 0.734217 | 0.550556 | -11.808083 | 0.509873 | 0.412025 | -12.166717 | 29.166963 | 3.406747 | -8.139884 | 31.624467 | 3.485063 | -8.058990 | 1.945226 | 1.080185 | -10.847469 | 2.085520 | 1.126720 | -10.777834 | 0.583333 | 0.583333 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 |
| 4 | 1013 | 1013 | Alabama | 18 | 1013 | greenvilleadvocate-greenvilleadvocate.com | -86.617752 | 31.829597 | The Greenville Advocate | 1510346.0 | 3 | 1 | 3 | 224 | 6 | 120 | 45 | 104 | 75 | 2 | 1 | 4 | 1 | 10464 | 436 | 8438 | 6752 | 2026 | 1686 | 317 | 245 | 119 | 72 | USA | 1369338 | 82081 | 1203928 | 1012453 | 165410 | 191475 | 70834 | 58064 | 11247 | 12770 | 2020-05-06 | Butler | 4901 | 3726 | 20280 | 52.781065 | 43.515779 | 1.247535 | 18.126233 | 18.940426 | 71.232157 | 6 | 9806370 | 19448 | 656479046 | 0.274104 | 534.759358 | 6.283685 | -5.221591 | 385.643768 | 5.957504 | -5.544818 | 20.567668 | 3.071195 | -8.266113 | 5.141917 | 1.815137 | -9.182404 | 20.660040 | 3.075469 | -8.484231 | 17.192906 | 2.901032 | -8.667836 | 1.213497 | 0.794574 | -11.311051 | 0.734217 | 0.550556 | -11.808083 | 25.196539 | 3.265627 | -8.286213 | 29.166963 | 3.406747 | -8.139884 | 1.713231 | 0.998140 | -10.974456 | 1.945226 | 1.080185 | -10.847469 | 1.000000 | 1.000000 | 22.32 | 31.51 | 46.16 | 19642 | 9.885425 | -14.227849 | 1013 | 3953 | 5448 | 0.320774 |
px.scatter(covid_df.sample(10000),
x='fips_n_cases_per1k_log',
y='log_odds_of_covid_article',
opacity=.5
)
px.scatter(covid_df.sample(10000),
x='country_n_deaths_per1k',
y='log_odds_of_covid_article',
opacity=.5
)
from scipy.stats import pearsonr
pearsonr(covid_df.country_n_deaths_per1k, covid_df.log_odds_of_covid_article )
(-0.04869514360602452, 1.5869820562104472e-08)
pearsonr(covid_df.fips_n_deaths_per1k, covid_df.log_odds_of_covid_article )
(-0.022215429530362464, 0.009958701688477182)
averaged_data = covid_df.groupby("date").agg(
cases = pd.NamedAgg("country_n_cases_per1k","first"),
deaths = pd.NamedAgg("country_n_deaths_per1k","first"),
article_odds = pd.NamedAgg("log_odds_of_covid_article","mean")
).reset_index()
fig = px.line(pd.melt(averaged_data,id_vars="date"),x="date",y="value",facet_col="variable")
fig.update_yaxes(matches=None)
fig.show()
Linear regression. Now we know what that is! And how to optimize it (although we'll use the sklearn implementation
Next week, we'll cover model evaluation in more detail. For now, we're just going to note that to ensure our model is generalizable and not overfit to the training data, we need to separate out a training dataset from a test dataset.
Exercise: Give a high-level argument for why evaluating on the training data is a bad idea
With temporal data, and more generally, data with dependencies, it is also important that we make sure that we are avoiding the leakage of information from the training data in a way that creates a biased understanding of how well we are making predictions. Leakage can happen in at least two ways (again, we'll go into more detail next week):
For this simple example, for now, we're going to ignore the leakage issue. We'll come back and fix that next week
Neat! But to pick a good training dataset, we first need to know ... what our dataset is. This data has a lot of features. In class, we'll play with a bunch of them, together. Here, I'm just going to get us started
CONTINUOUS_FEATURES = ["state_lag1_n_cases_per1k",
"fips_lag1_n_cases_per1k",
"country_lag1_n_cases_per1k",
"predrt_0"]
OUTCOME = 'log_odds_of_covid_article'
X = covid_df[CONTINUOUS_FEATURES]
y = covid_df[OUTCOME]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state=5)
OK, let's have at it!
from sklearn.linear_model import LinearRegression
model = LinearRegression()
model.fit(X_train,y_train)
LinearRegression()
predictions = model.predict(X_test)
from sklearn.metrics import mean_squared_error
mean_squared_error(y_test,predictions)
0.6643640375053314
What might we be asking ourselves before we deploy? What might we try to change? Let's work on it!
In class exercise ... beat Kenny's predictive model!
We have to be really careful when interpreting coefficients for models with transformed predictor variables. Here, for example, is a useful resource for your programming assignment.
In our case, we actually ended up using a variable that means interpreting our coefficients using interpretations for logistic regression. Here is a good explanation. We will cover this in more detail next week, but a simple plot below to discuss!
Exercise: Are the estimates from the coefficients we used comparable? Which are, and which are not? What might we do to make them even more comparable?
For this demo, I took code from this sklearn tutorial. The tutorial is very nice, I would highly recommend going through it, although I will teach most of what is in this over the next week or two in one way or another.
feature_names = CONTINUOUS_FEATURES
coefs = pd.DataFrame(
model.coef_,
columns=["Coefficients"],
index=feature_names,
)
coefs
| Coefficients | |
|---|---|
| state_lag1_n_cases_per1k | 0.000557 |
| fips_lag1_n_cases_per1k | -0.000172 |
| country_lag1_n_cases_per1k | -0.002770 |
| predrt_0 | -0.013255 |
import matplotlib.pyplot as plt
coefs.plot(kind="barh", figsize=(9, 7))
plt.title("Ridge model, small regularization")
plt.axvline(x=0, color=".5")
plt.subplots_adjust(left=0.3)
coefs['odds_ratio'] = np.exp(coefs.Coefficients)
coefs
| Coefficients | odds_ratio | |
|---|---|---|
| state_lag1_n_cases_per1k | 0.000557 | 1.000557 |
| fips_lag1_n_cases_per1k | -0.000172 | 0.999828 |
| country_lag1_n_cases_per1k | -0.002770 | 0.997234 |
| predrt_0 | -0.013255 | 0.986832 |